|
||
|
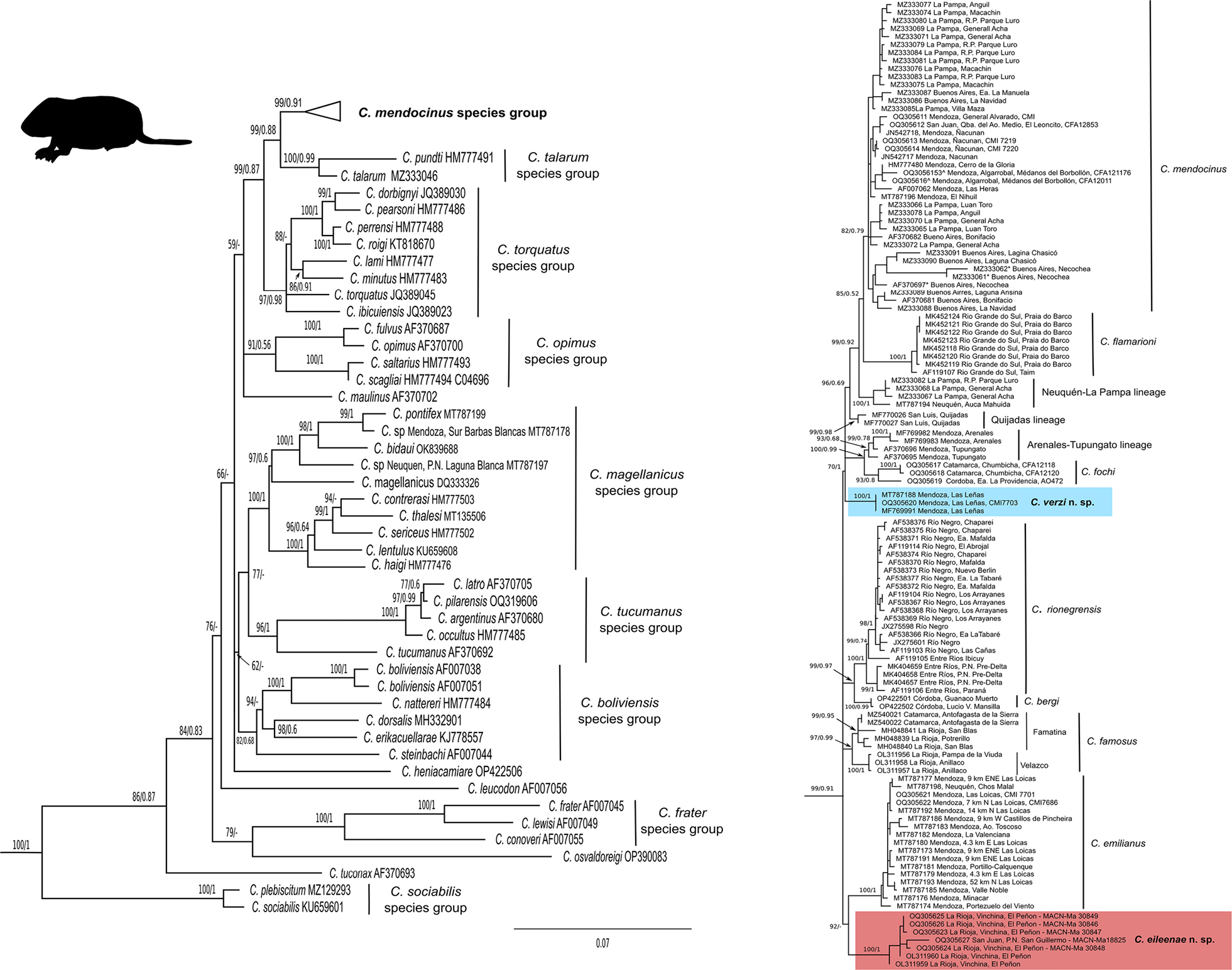
Maximun Likelihood tree (ln = –12720.876) obtained in the analysis of 164 cytochrome b gene sequences of the Ctenomys. Numbers indicate bootstrap (right of the diagonal) and posterior probability (left of the diagonal) values of adjacent nodes; a missing value indicates that the given node has less than 50% of posterior probability; a dash indicates that the given node was not recovered in the Bayesian analysis. For clarity the tree is split into two. In the left panel the general tree of Ctenomys is shown (outgroup not shown); species groups (sensu D’Elía et al. 2021) are indicated with bars (the two main clades of C. famosus are also labeled as in Tammone et al. 2022a). In the right panel the focus is on the Ctenomys mendocinus species group. Terminal labels indicate species name and GenBank accession numbers. For the species of the Ctenomys mendocinus species group, terminal labels include Province/State/Department and abbreviated locality (see more details in Supplementary File 1). The silhouette of Ctenomys was taken from PhyloPic and was made available by Chloé Schmidt under a license CC BY 3.0. |