|
||
|
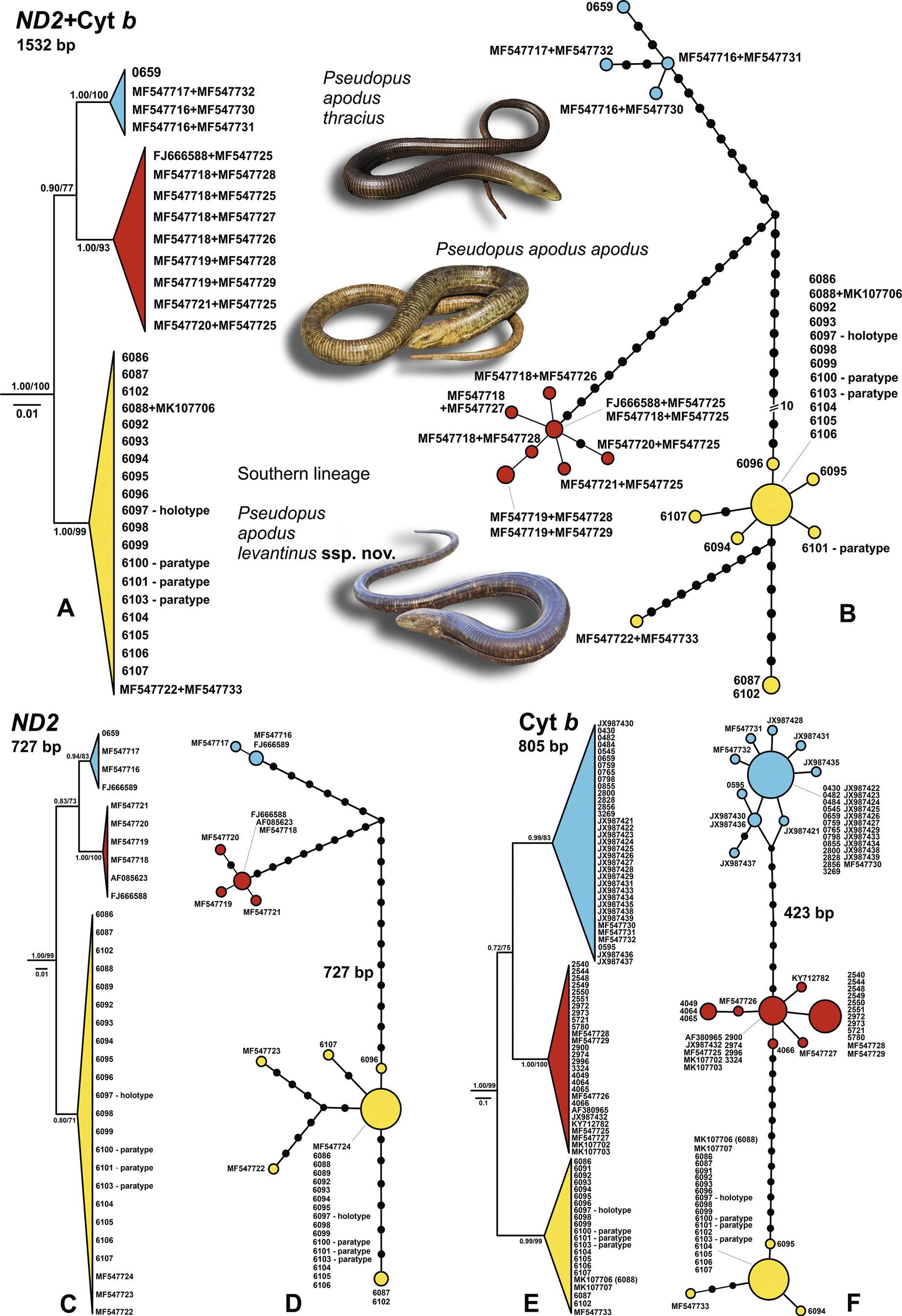
Phylogenetic relationships of Pseudopus apodus reconstructed from concatenated and single locus trees of ND2 & Cyt b sequences and presented as collapsed Bayesian trees (A, C, E) and haplotype networks (B, D, F). The numbers at the tree nodes represent Bayesian Posterior Probabilities/Maximum Likelihood Bootstraps showing branch support (for details see Figures S1 and S2). The numbers at the end of main lineages or with haplotypes show the sampling code (DJab acronym) or GenBank numbers (see also Table S1). Circle sizes in the haplotype networks are proportional to the relative frequency of the haplotypes and small black circles represent the missing haplotypes. |