|
||
|
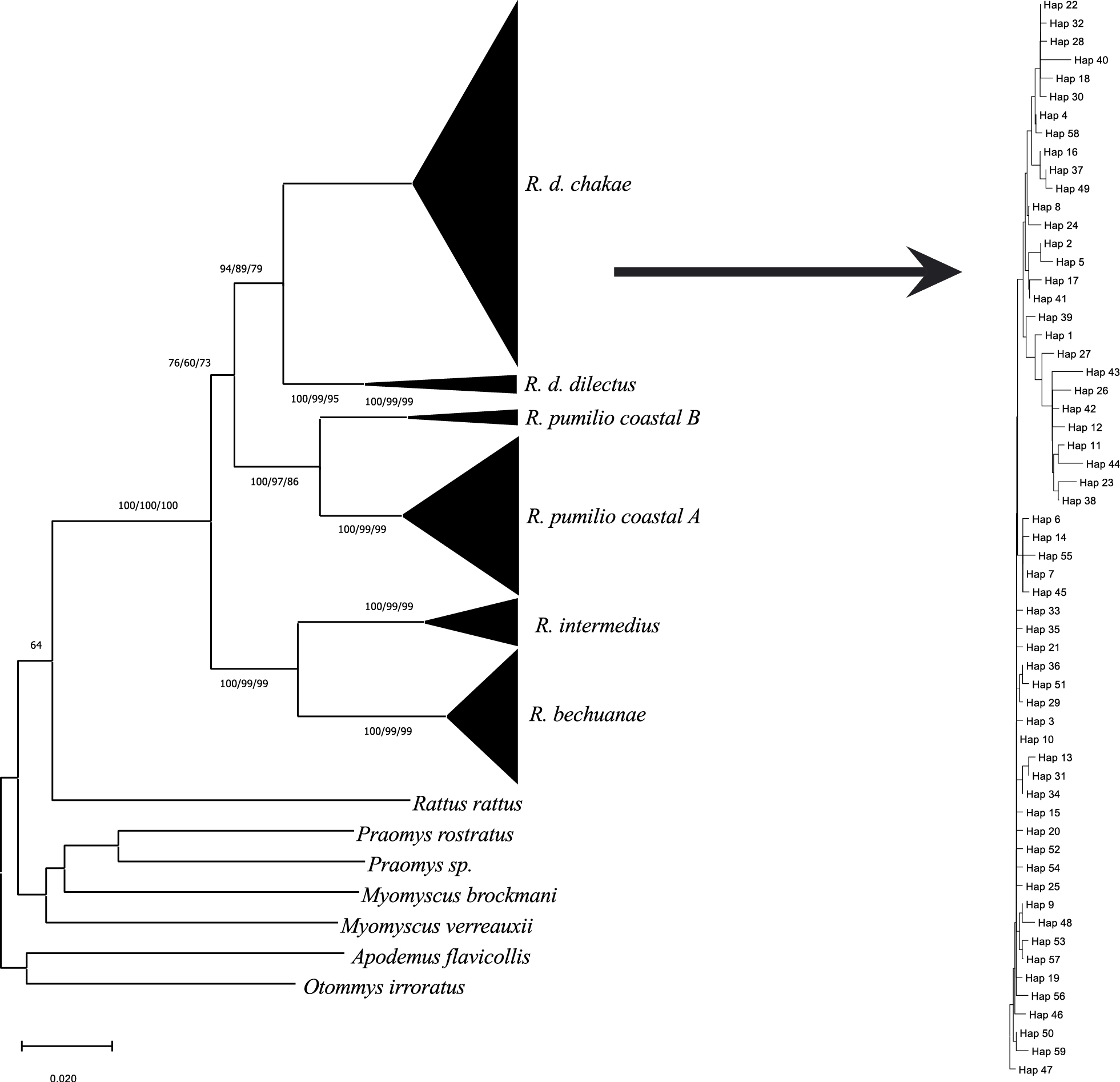
A Neighbour-joining (NJ) phylogram illustrating phylogenetic relationships among haplotypes generated from a total of 280 Rhabdomys COI sequences. The dataset includes 101 sequences from this study with other Rhabdomys sequences downloaded from GenBank. The included outgroups were two sequences of Praomys species and Myomyscus species, and a single sequence of Rattus rattus, Apodemus flavicollis and Otomys irroratus. The tree is condensed, while the insert shows R. d. chakae which includes all samples from this study. The Neighbour-joining Maximum Likelihood (ML), and Maximum Parsimony (MP) analyses were drawn using the Kimura 2-Parameter model (Kimura 1980). Bootstrap resampling support (1000 iterations) are shown for uncut nodes, respectively. Evolutionary analyses were conducted in MEGA version X (Kumar et al. 2018). |