|
||
|
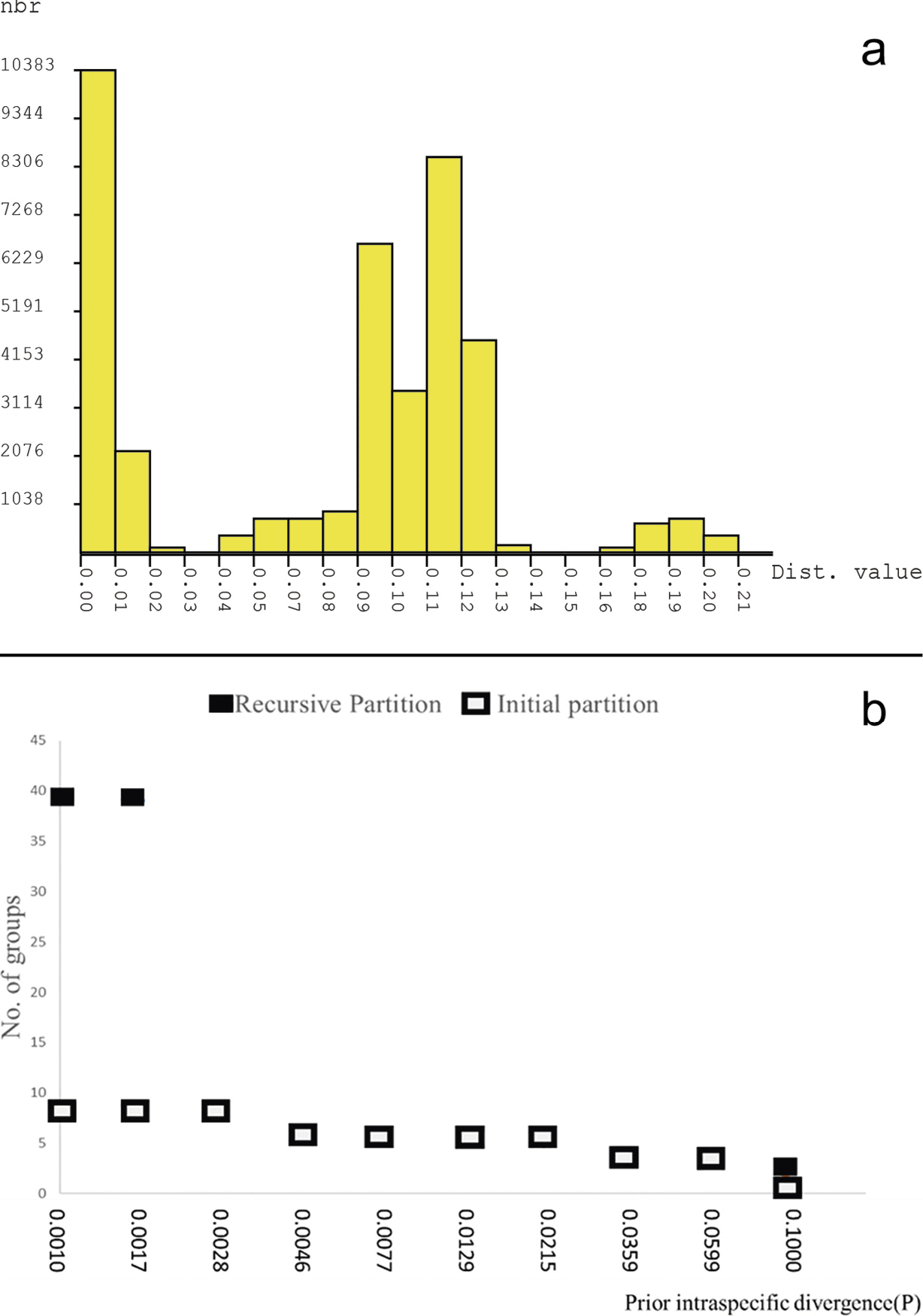
Barcode gap analysis of Rhabdomys taxa generated by Automatic Barcode Discovery Gap Discovery (Puillandre et al. 2012) using 280 sequences in the COI gene (582 bp). The gap was calculated with the default setting using Kimura (K80) TS/TV distances. (a) Histograms showing the frequency of pairwise nucleotide differences, and (b) shows a plot with the number of PSHs (preliminary species hypothesis) obtained for each prior intraspecific divergence (0.01–0.1000). |