|
||
|
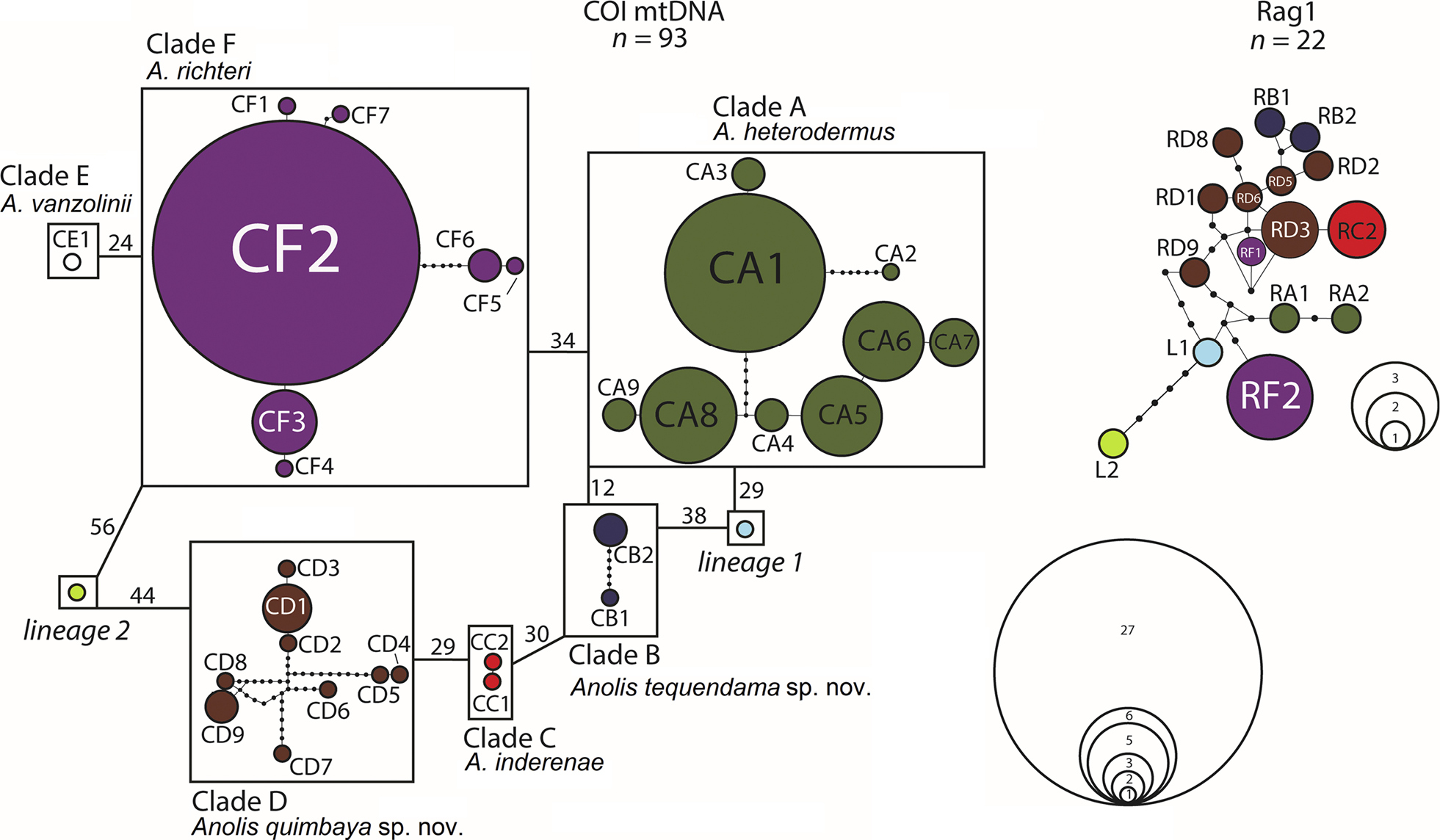
Parsimony networks for mitochondrial (left) and nuclear (right) haplotypes of the heterodermus subgroup (sensu Williams et al. 1996), based on alignments of 1092 bp of mtDNA and 1057 bp of the intron 1 of the nuclear R35 gene (n = number of individuals). Colour code as in Figs 1 and 2. Circle size reflects haplotype frequency; missing haplotypes are represented by small black circles. Each line connecting haplotypes corresponds to one mutational step. Mitochondrial haplotypes or haplotype clusters shown in boxes remain unconnected under the 90%–95% thresholds. Numbers next to lines linking clusters indicate minimum fixed differences between clusters. Connection limit for nuclear haplotypes is 95%. The colours of haplotypes correspond to respective mitochondrial haplotypes. For abbreviations, see Figure 2. |