|
||
|
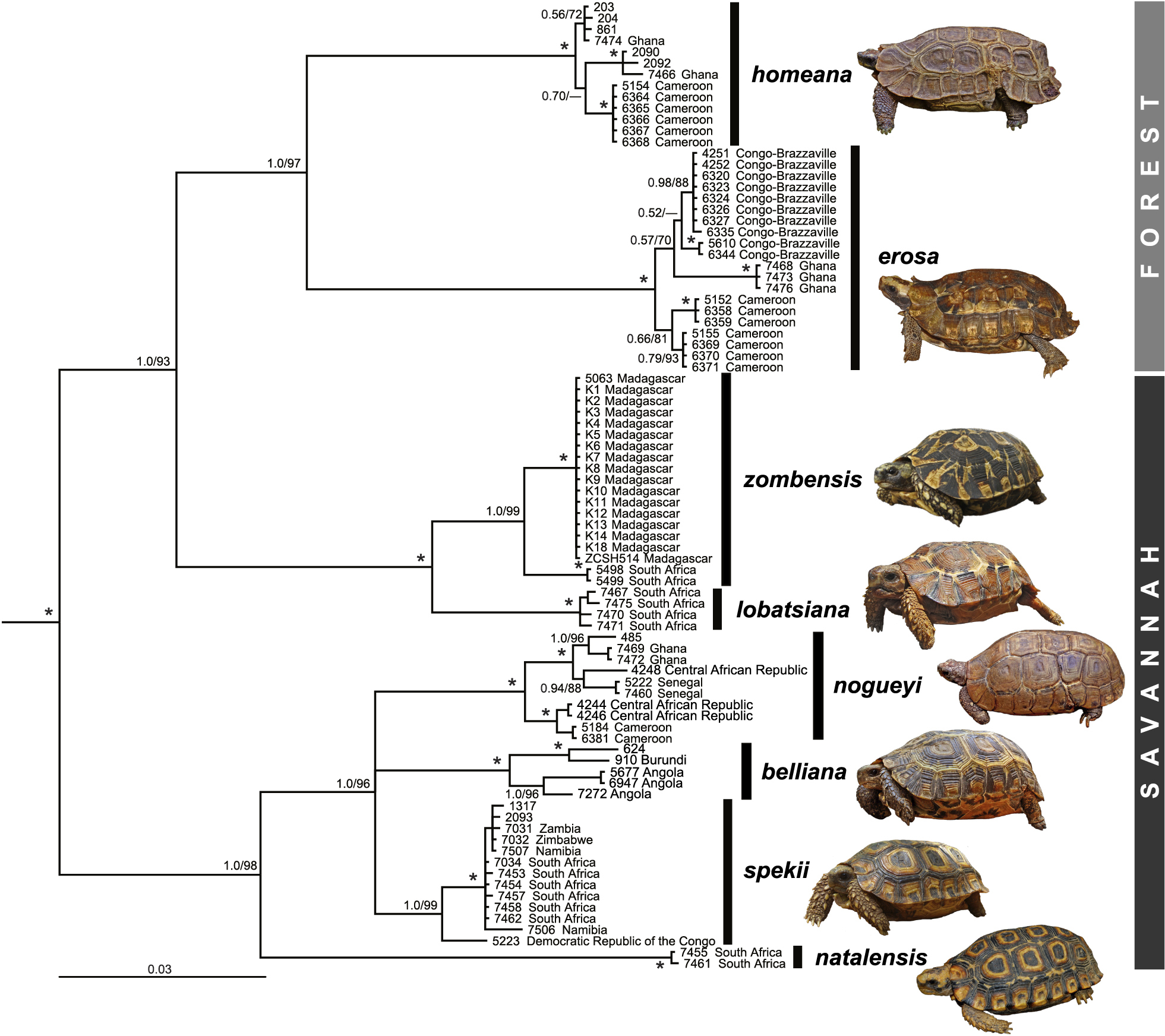
Mitochondrial phylogeny of Kinixys species based on a 2,273-bp-long alignment of mtDNA (Bayesian analysis). Numbers at nodes are posterior probabilities and bootstrap values from a maximum likelihood analysis; asterisks indicate maximum support under both approaches (redrawn from Kindler et al. 2012; for sample codes, see there). For the widely distributed species (Kinixys homeana, K. erosa, K. zombensis, K. nogueyi, K. belliana, K. spekii), the internal branching structure shows phylogeographic variation is present in the clades. Insets: K. homeana, pet trade (photo: Pavel Široký); K. erosa, Democratic Republic of the Congo (photo: Václav Gvoždík); K. zombensis, South Africa (photo: Flora Ihlow); K. lobatsiana, South Africa (photo: Flora Ihlow); K. nogueyi, pet trade (photo: Pavel Široký); K. belliana, Angola (photo: Luis Ceríaco); K. spekii, South Africa (photo: Flora Ihlow); K. natalensis, South Africa (photo: James Harvey). |