|
||
|
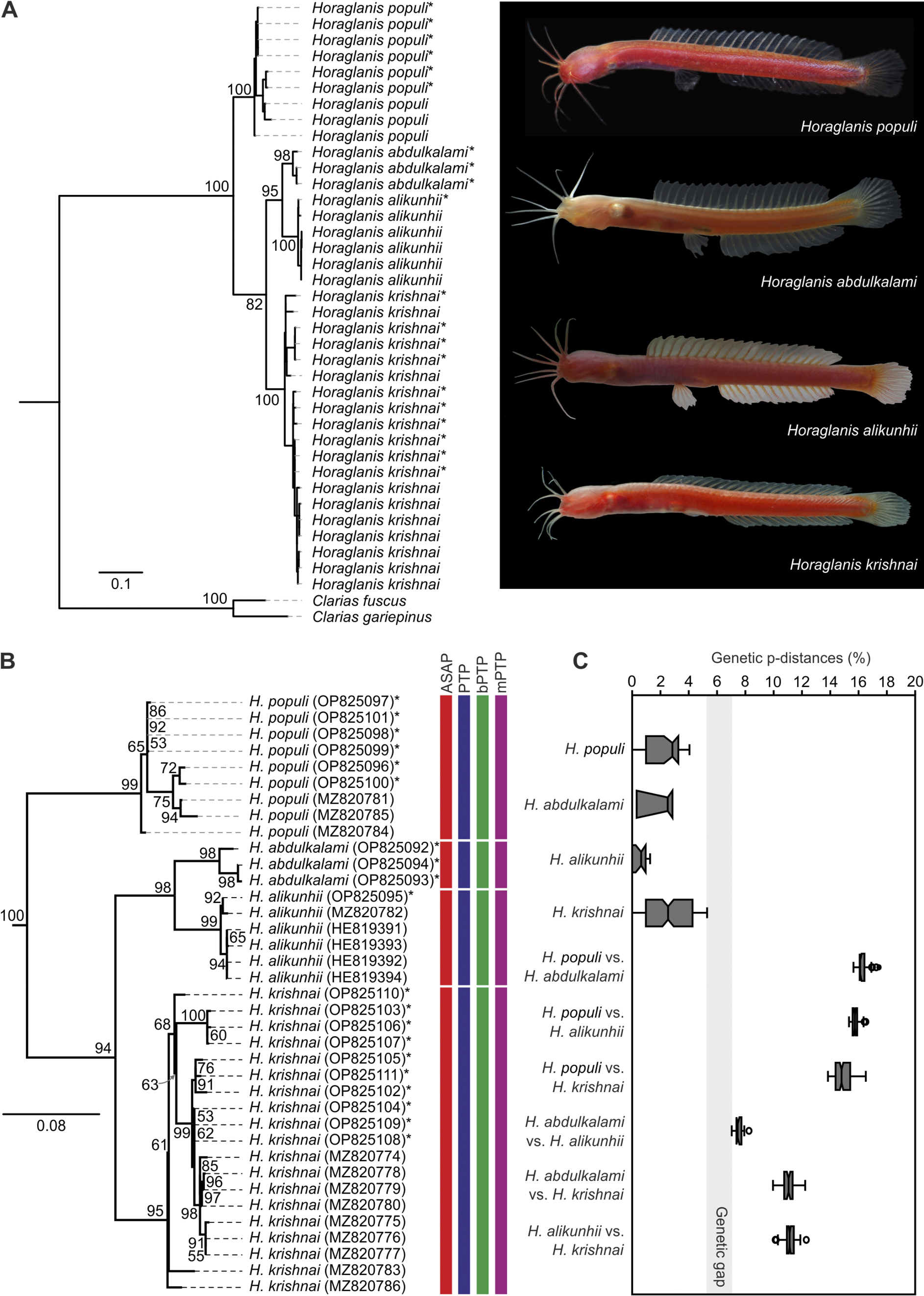
Phylogenetic tree of species of Horaglanis and their delimitation. A Maximum likelihood phylogenetic tree based on concatenated mitochondrial COI, cyt b, 12S rRNA and 16S rRNA gene sequences employing best partition scheme and nucleotide substitution models. B Maximum likelihood phylogenetic analysis based on COI gene employing best partition scheme and nucleotide substitution models. Species delimitation based on ASAP, PTP, bPTP and mPTP processes shown as bars adjacent to species names. C Box plots of intraspecific and interspecific genetic p-distances in COI gene. Genetic gap between greatest intraspecific (5.3%) and smallest interspecific (7.0%) genetic distance shown in light grey. A, B Clarias species used as outgroups. Values along nodes are bootstrap supports based on 1000 iterations. Asterisks indicate sequences generated in the current study. |